Great Lakes Bioinformatics
Conference (GLBIO)
May 20, 2019
Tutorial on Dimensionality Reduction Methods for Biomedical Data
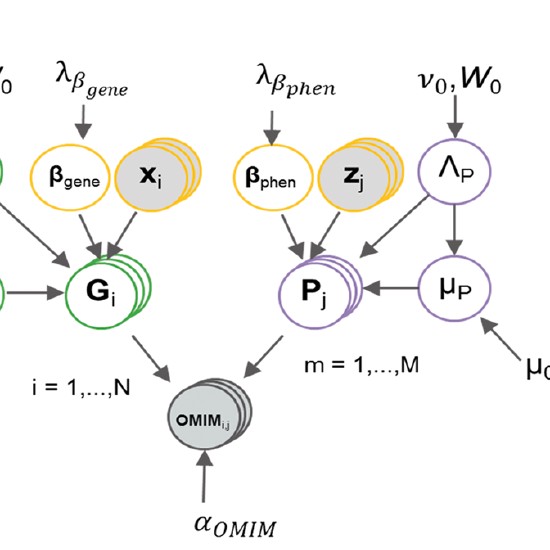
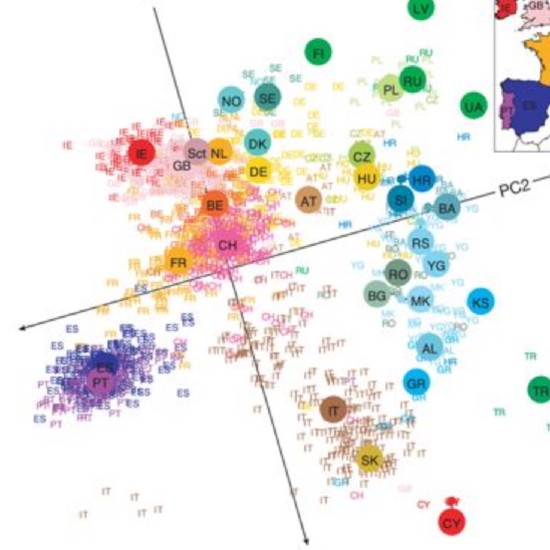
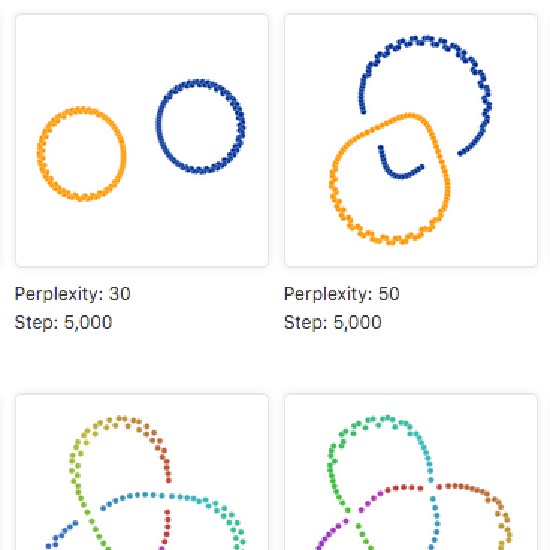
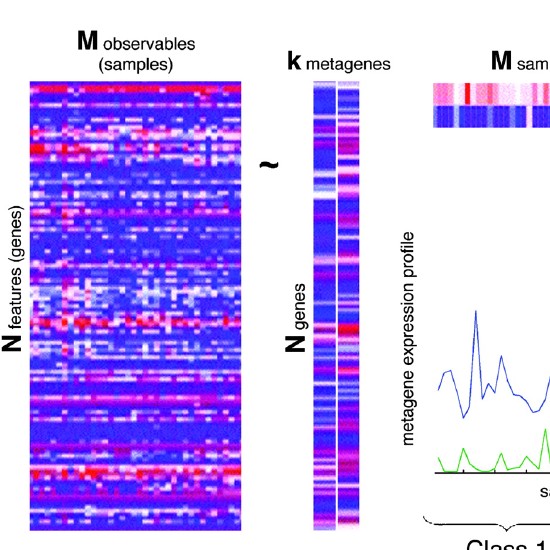
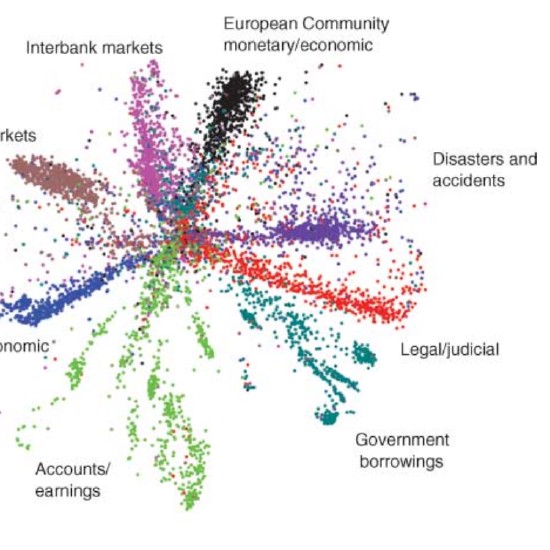
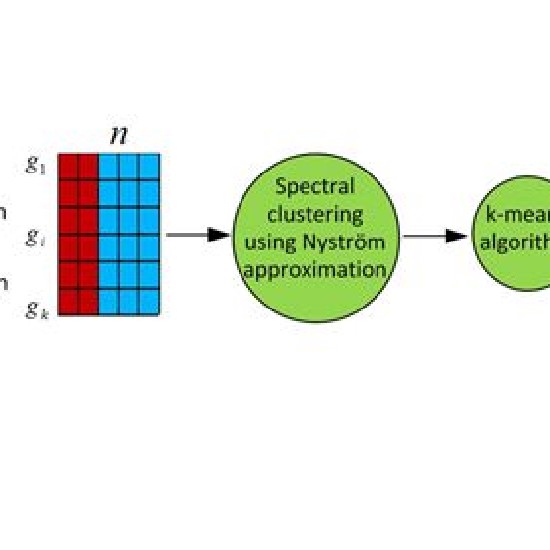
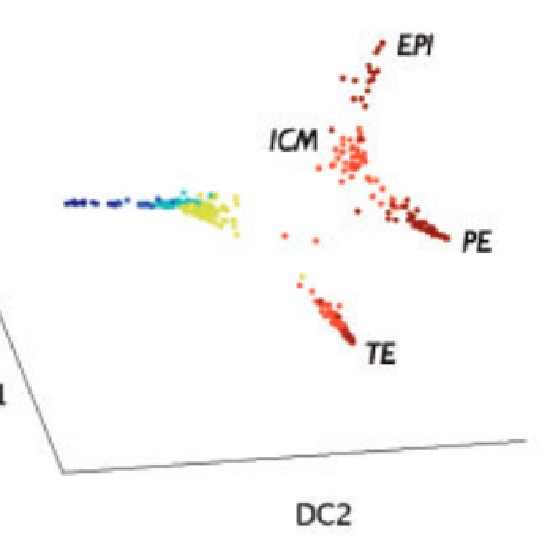
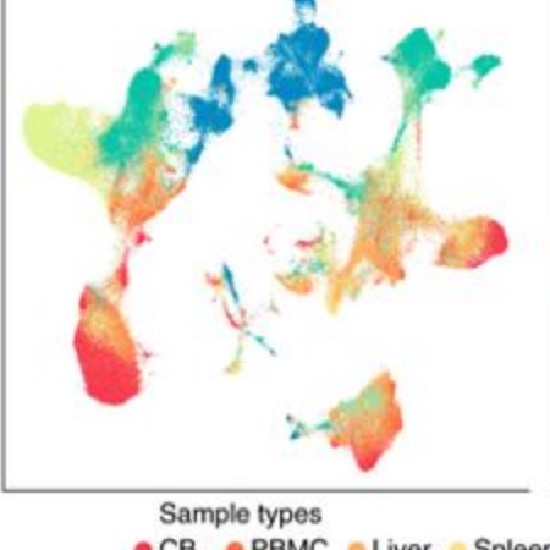
Many real-world datasets are high dimensional in their raw form but have low-dimensional structure, groupings, or representations. Dimensionality reduction methods have been applied to various biomedical datasets with the aim of cancer subtype extraction from mutational signatures, genotype-to-phenotype mapping, gene regulatory program identification, unsupervised multi-omics data integration, and cell differentiation trajectory visualization. This workshop will provide an opportunity to explore a handful of powerful dimensionality reduction methods: matrix factorization, PCA/LDA/GDA, t-SNE and UMAP, diffusion map, and autoencoders. All demos and exercises will use real biomedical datasets from single cell RNA-seq, Hi-C, and de-identified medical records.Topics
-
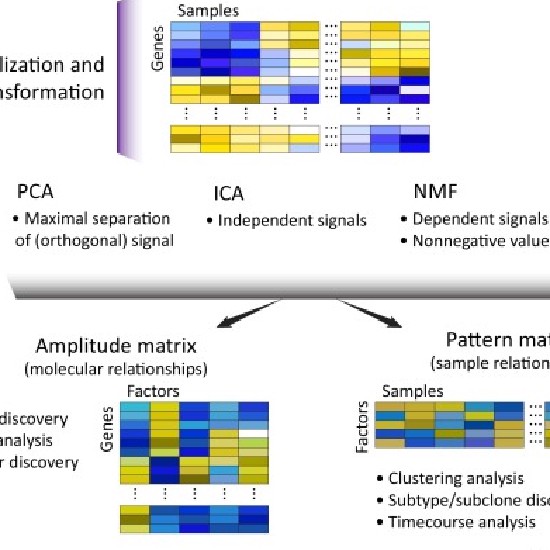
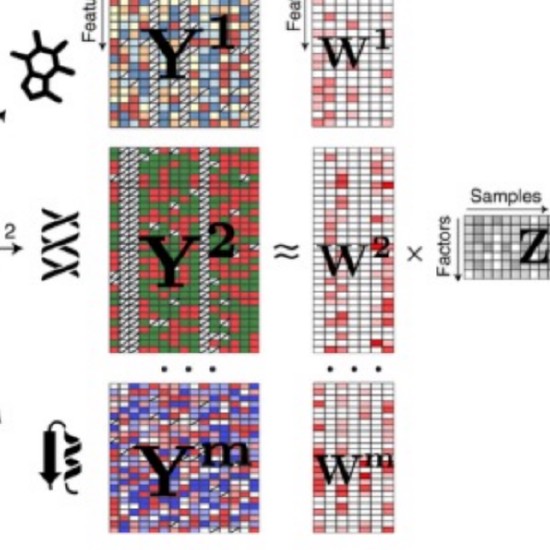
Non-negative Matrix Factorization (NMF)
-
Principal Component Analysis (PCA) & family
-
t-SNE and UMAP
-
Diffusion map
-
Autoencoders
Datasets
-
Single cell RNA-seq datasets
-
Hi-C datasets
-
Deidentified medical records